Among difficult-to-express proteins, membrane proteins stand out notoriously – unstable, low-yielding, and aggregation-prone – often unyielding to our best efforts to express them successfully. Yet, as well-established drug targets, contributing to around 55% of the biologics market (1), membrane protein expression is crucial in drug development pipelines. From target characterization and validation through hit identification, lead optimization, and ultimately selecting a candidate molecule for clinical development, optimizing their expression is critical for success.
Here, we explore some key reasons why your membrane protein expression might be failing and ways to tackle them.
1. Misfolding and aggregation in heterologous expression hosts
Membrane proteins have complex structures, making them unstable outside their native cell membrane environment and, thereby, highly challenging to produce in viable quantities. The go-to expression hosts for heterologous membrane protein expression, such as bacterial and yeast cells, may not correctly fold membrane proteins, leading to their misfolding and aggregation.
Tackle this by: mutating specific residues in your protein to reduce reliance on host-specific lipids or by fusing your protein to solubility tags such as superfolderGFP to increase stability.
2. Incorrect integration into the lipid bilayer
For optimal function, membrane proteins must be correctly integrated into the lipid bilayer. Proper folding and insertion into the membrane can be challenging, as the folding process often requires assistance from chaperones and other membrane proteins. Moreover, the lipid composition of the target membrane should be as close as possible to the native lipid environment of your target protein.
Tackle this by: adding chaperones and integrases to the expression reaction to assist with lipid bilayer insertion, by adjusting the lipid bilayer composition through lipid engineering (2), or by slowing down translation to allow more time for integration, such as by reducing the expression temperature in E. coli.

3. Detergent-induced destabilization of the protein structure
Membrane proteins possess a hydrophobic transmembrane segment, which renders them water-insoluble, necessitating the use of detergents for their extraction and purification. However, excessive use of detergents strips away structure-stabilizing lipids away from proteins, leading to aggregation or denaturation of proteins.
Tackle this by: evaluating various detergent types and concentrations to determine the right detergent and optimizing the detergent-to-lipid ratio. Mixing the detergent with cholesterol homologs (CHS) to mimic fluidity (e.g., human lipid bilayers) can also help.
4. Loss of protein function in a non-native environment
Retaining the functionality and activity of membrane proteins after extraction from their native environment is often the biggest challenge with membrane protein extraction. Some membrane proteins may lose function outside the lipid bilayer and their specific cellular environment or display altered activity.
Tackle this by: reintegrating the isolated protein into artificial bilayer systems like Nano discs.
Already tried all the tips and tricks and still can’t get your protein to express how it should? It might be time to think ‘outside the cell.’
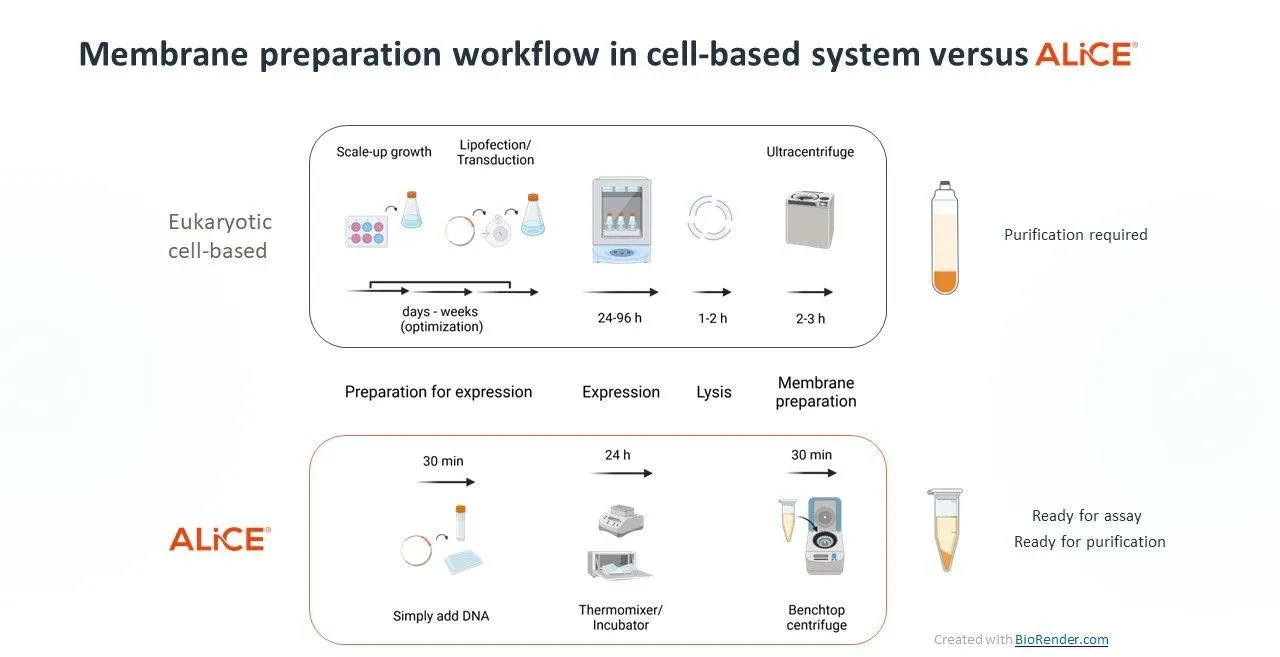
Cell-free protein expression (CFPE) harnesses the cell’s protein synthesis machinery to express a myriad of proteins, including difficult-to-express membrane proteins, in high quantities.
Eukaryotic cell-free systems like our plant-based platform, ALiCE®, overcome many of the challenges of membrane protein expression without extensive optimization. ALiCE® contains native microsomes derived from the endoplasmic reticulum and Golgi, which form a scaffold into which membrane proteins can insert themselves. The ability of ALiCE® to reliably produce functional membrane proteins makes it a viable and faster alternative to cell-based expression.
Intrigued? Download our application note to explore rapid membrane protein expression in ALiCE®
1. Sood, Vanita D., Alec W. Gross. Membrane proteins as targets for biological drugs. Advances in membrane proteins: building, signaling, and malfunction (2019): 49–65.
2. Pichler, Harald, and Emmerstorfer-Augustin, Anita. Modification of membrane lipid compositions in single-celled organisms – From basics to applications.” Methods (San Diego, Calif.) vol. 147 (2018): 50–65.